Research Interests
My own research investigates a wide variety of topics within the realm of computational ecology and imaging biology. My current focus is predominantly on spider web imaging and web traits.
My work is largely theoretical and computational, but also includes lab and fieldwork as the empirical foundation. I look to improve current methods of data generation, analysis, and visualisation, incorporating my experience as a graphic designer and in software development.
Novel imaging methods
Imaging concepts and techniques are important in ecology and in biology more generally. Camera trapping, camouflage, biomimicry, vision, colouration, and structural analysis all have major elements of image generation and analysis. I am very interested in designing and testing new methods to extract this sort of data in the field and the lab, with a current focus on spider web imaging. In my recent paper "Improving in-situ spider web photography with UV fluorescence" (in review), I developed a modification to the established night-sampling techniques used to photograph spider webs in the field. I exploited the phenomenon of UV fluorescence to force silk strands to glow in the visible light spectrum while excluding any UV light reflected from the background with a UV filter. This notably improved contrast within photos, leading to much better distinction between webs and their backgrounds.

Image analysis pipelines
I usually take a mechanistic approach to image analysis: I start from the physical fundamentals of how the features of an image are created and use that knowledge to design analyses that extract the traits we are interested in. Part of my current work in my PhD focuses on automating the process of extracting functional traits from images of spider webs. This has historically been a slow and manual process, but our methods are around 52x faster and create digitisation outputs that are more data-rich.
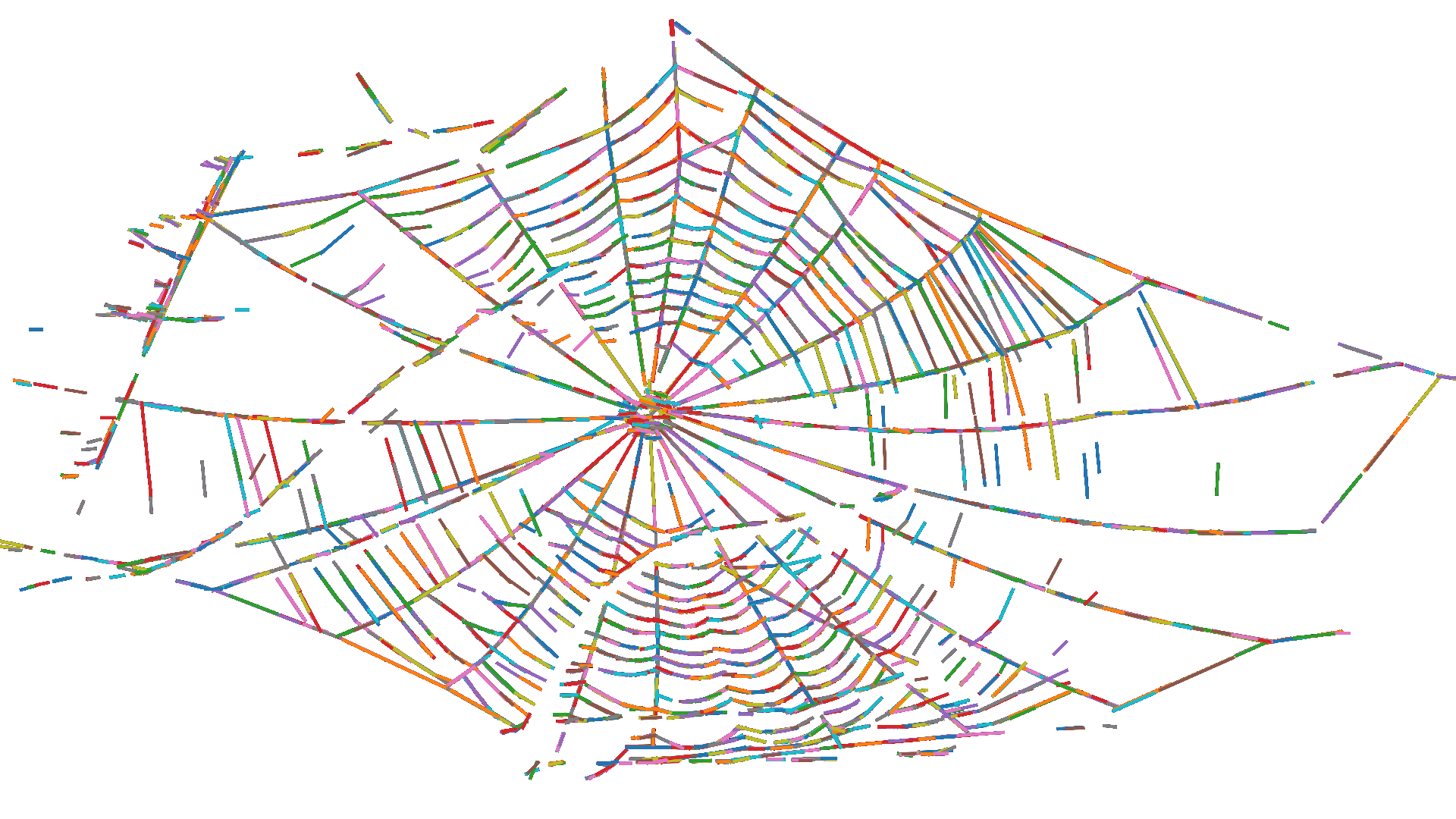
Computational ecology methods
Ecology benefits hugely from accurate and well-designed computing. I focus on improving the speed and reproducibility of ecological analysis and modelling using powerful, extensible, user-friendly computational approaches. This aim also filters down to my teaching, where I work to empower students to both write clever code and also to believe that they can solve problems in computing using their own intuition and skill. I predominantly work in Python and R, but also write in C, C++, Javascript, and Processing.
Spider web traits
My chosen system of study at present is that of spider web traits. Orb-weaver spiders change the structure of their webs based upon both internal and external conditions. By measuring these webs we can gain insight into the conditions under which the spiders built. Spiders are historically understudied creatures, however given their roles as generalist predators they are also critical to the function of terrestrial ecosystems. I look to improve the data sources that we have about spider web traits, the methodologies for collecting these data, and to use these to interrogate questions of scaling and temperature dependence in spider foraging.

Other research interests
My current position is focused predominantly on centralising and increasing access to population, trait, and genomic data for researchers working on modelling disease vectors in the UK. This includes web-based and programmatic tools to assist in data access, processing, and cross-linking. I will also be designing a suite of interactive visualisations of this data to aid policy-makers and scientists alike in making decisions on disease vector-related issues.



